How To Align Protein Sequences . The program compares nucleotide or protein. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. Then use the blast button at the. this episode of the blast tutorial series demonstrates how to compare or align two. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. the basic local alignment search tool (blast) finds regions of local similarity between sequences. enter one or more queries in the top text box and one or more subject sequences in the lower text box. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):.
from medium.com
select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. this episode of the blast tutorial series demonstrates how to compare or align two. the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. enter one or more queries in the top text box and one or more subject sequences in the lower text box. Then use the blast button at the. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a.
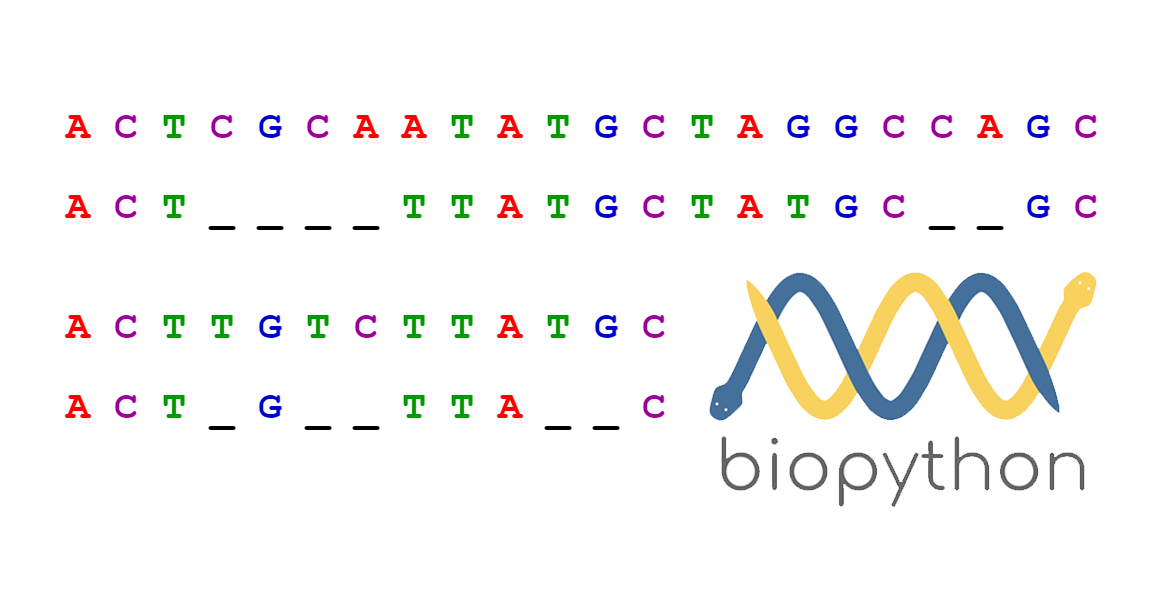
Pairwise Sequence Alignment using Biopython Towards Data Science
How To Align Protein Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. The program compares nucleotide or protein. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. Then use the blast button at the. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. enter one or more queries in the top text box and one or more subject sequences in the lower text box. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. this episode of the blast tutorial series demonstrates how to compare or align two. the basic local alignment search tool (blast) finds regions of local similarity between sequences.
From www.researchgate.net
Showing multiple sequence alignment of protein sequence of Subunit A How To Align Protein Sequences select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. the basic local alignment search tool (blast) finds regions of. How To Align Protein Sequences.
From www.researchgate.net
Multiple sequence alignment of the protein sequence SmCL1 and other six How To Align Protein Sequences we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. enter one or more queries in the top text box and one or more subject sequences in the lower text box. The program compares nucleotide or protein. Then use the blast button at the. the basic local alignment search tool (blast) finds. How To Align Protein Sequences.
From www.researchgate.net
Protein sequence alignment of keratinase and other proteases by ESPript How To Align Protein Sequences a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. this episode of the blast tutorial series demonstrates how to compare or align two. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. Then use the blast button. How To Align Protein Sequences.
From www.youtube.com
How to align Protein sequences relationship UniProt 2 How To Align Protein Sequences The program compares nucleotide or protein. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. Then use the blast button at the. this episode of the blast tutorial series demonstrates how to compare or align two. a sequence alignment is a way of arranging the primary sequences of a protein to. How To Align Protein Sequences.
From www.researchgate.net
Multiple alignment of protein sequence and phylogeny of the four cloned How To Align Protein Sequences a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. The program compares nucleotide or protein. this episode of the blast tutorial series demonstrates how to compare or align two. the basic local alignment search tool (blast) finds regions of local similarity between sequences.. How To Align Protein Sequences.
From www.researchgate.net
Amino acid sequence alignment of cSOD1 with seven different mammalian How To Align Protein Sequences we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. the basic local alignment search tool (blast) finds regions of local similarity between sequences. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. The program compares nucleotide. How To Align Protein Sequences.
From www.researchgate.net
Alignment of the Protein Sequences of the Eight Known CDPSs and the How To Align Protein Sequences this episode of the blast tutorial series demonstrates how to compare or align two. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf. How To Align Protein Sequences.
From www.youtube.com
How to Use BLAST for Finding and Aligning DNA or Protein Sequences How To Align Protein Sequences select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. this episode of the blast tutorial series demonstrates how to compare or align two. The program compares nucleotide or protein. enter one or more queries in the top text box and one or. How To Align Protein Sequences.
From www.researchgate.net
Protein sequence and structure alignment of JAK family PTK domain. (A How To Align Protein Sequences a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. The program compares nucleotide or protein. this episode of the. How To Align Protein Sequences.
From www.researchgate.net
Comparison of the protein sequences from 79 StWRKY proteins. Alignment How To Align Protein Sequences this episode of the blast tutorial series demonstrates how to compare or align two. Then use the blast button at the. The program compares nucleotide or protein. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. select the align tab of the toolbar. How To Align Protein Sequences.
From medium.com
Pairwise Sequence Alignment using Biopython Towards Data Science How To Align Protein Sequences select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. The program compares nucleotide or protein. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. this episode of the blast tutorial series demonstrates how to compare or. How To Align Protein Sequences.
From www.researchgate.net
Protein sequence alignment of A. ricciae GH45 endoglucanases. Orange How To Align Protein Sequences this episode of the blast tutorial series demonstrates how to compare or align two. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. the basic local alignment search tool (blast) finds regions of local similarity between sequences. select the align tab of. How To Align Protein Sequences.
From www.researchgate.net
Multiple amino acid sequence alignment of 4/1like proteins. The amino How To Align Protein Sequences the basic local alignment search tool (blast) finds regions of local similarity between sequences. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf. How To Align Protein Sequences.
From www.creative-biostructure.com
Protein Sequence Analysis for Coronavirus Research Creative How To Align Protein Sequences we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. this episode of the blast tutorial series demonstrates how to compare or align two. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. Then use the blast button. How To Align Protein Sequences.
From www.biologyexams4u.com
Importance of Sequence Alignment in Bioinformatics Biology Exams 4 U How To Align Protein Sequences this episode of the blast tutorial series demonstrates how to compare or align two. we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between sequences. enter one or more queries in the. How To Align Protein Sequences.
From www.ucl.ac.uk
Multiple sequence alignment How To Align Protein Sequences we introduce dedal, a trainable algorithm for accurate pairwise local alignment of protein sequences (fig. Then use the blast button at the. The program compares nucleotide or protein. enter one or more queries in the top text box and one or more subject sequences in the lower text box. select the align tab of the toolbar to. How To Align Protein Sequences.
From www.researchgate.net
Multiple sequence alignment of the MuPR1 protein sequence with other How To Align Protein Sequences The program compares nucleotide or protein. select the align tab of the toolbar to align two or more protein sequences with the clustal omega program (cf also this clustalo faq):. a sequence alignment is a way of arranging the primary sequences of a protein to identify regions of similarity that may be a. enter one or more. How To Align Protein Sequences.
From www.researchgate.net
Multiple protein sequence alignment of HTT exon 29. Multiple protein How To Align Protein Sequences this episode of the blast tutorial series demonstrates how to compare or align two. The program compares nucleotide or protein. the basic local alignment search tool (blast) finds regions of local similarity between sequences. enter one or more queries in the top text box and one or more subject sequences in the lower text box. select. How To Align Protein Sequences.